Python alongside R
Nikita Gusarov
While working on my PhD thesis I’m usually facing the situation when I need to use both R and Python programming languages.
R offers me the possibility to render neat scientific supports with rmarkdown package, which bring .Rmd support.
Obviously, Python has similar format (pymarkdown and the .Pmd) available, it is not as well developed as the R counterpart.
Moreover, I’m far more familiar with R statistical libraries, tidyverse suite and coding practices.
However, at the same time I need Python dependencies: the most developed neural network (NN) libraries are written in Python or have their main interface in Python.
For example, the well known TensorFlow has a Python based backend.
The idea behind this post is to describe the possibilities to combine both R and Python in your workflow. There is quite a lot of disjoined materials available on internet in this post, leaving not that much place for my personal contribution. But I hope it might be useful for someone, and I expect to convert the results into a guide for the TER’s and other courses I’m giving.
Configuring reticulate
One of the most developed packages for R and Python interactions is the rticulate.
It offers several possible strategies of interaction between the languages, among which:
- Calling Python directly from R
- Sourcing Python scripts
- Including Python chunks in RMarkdown
- Using Python directly from within an R session
- Translation between R and Python objects
- Interactions between the different versions of Python through virtual environments
The reticulate is provided as CRAN package to be installed with R:
# Install reticulate
install.packages("reticulate")The package uses the Python version found on your PATH by default.
So if you already have a working Python installation there is nothing to do.
However, given the Python language specifics I suggest you use a separate environment to use alongside your R installation.
One of the good practices is to use a Conda virtual environment, or Python virtualenv.
If you desire to configure a separate Python installation, the reticulate package offers such possibility via miniconda installation.
This may be achieved with a simple command:
# Install miniconda
reticulate::install_miniconda()The miniconda will be installed in the default, system specific, path.
To modify it, one can set environmental variable RETICULATE_MINICONDA_PATH.
After the installation do not forget to verify that conda is in your PATH and activated.
An existing Python installation can provide similar experience through virtualenv functionality.
To create a new virtual environment execute:
# Call reticualte
library(reticulate)# Create new virtual environment
virtualenv_create("r-environemnt")
# Tell reticulate what to use
use_virtualenv("r-environment")To ensure the correct detections of the desired default Python installations by reticulate you may want to define RETICULATE_PYTHON environmental variable.
Finally, you may verify the activated environment’s configuration with:
# Configuration info
py_config()## python: /home/nikita/.local/share/r-miniconda/envs/r-reticulate/bin/python
## libpython: /home/nikita/.local/share/r-miniconda/envs/r-reticulate/lib/libpython3.7m.so
## pythonhome: /home/nikita/.local/share/r-miniconda/envs/r-reticulate:/home/nikita/.local/share/r-miniconda/envs/r-reticulate
## version: 3.7.10 | packaged by conda-forge | (default, Oct 13 2021, 21:01:18) [GCC 9.4.0]
## numpy: /home/nikita/.local/share/r-miniconda/envs/r-reticulate/lib/python3.7/site-packages/numpy
## numpy_version: 1.21.2Once the Python is setup and reticulate detects the desired environment, we may proceed to the description of usecases.
Interacting with Python
As you could see, there exist multiple strategies to interact with Python from inside R.
In order to better illustrate how exactly the interaction between R and Python is performed it’s better to start with REPL path.
Calling Python REPL from within R
This first option allows us to switch between R and Python sessions in terminal and access shared objects.
For those who do not know what REPL is here is a short definition:
A read–eval–print loop (REPL), also termed an interactive toplevel or language shell, is a simple interactive computer programming environment that takes single user inputs, executes them, and returns the result to the user …
We can access Python REPL interface directly from R by running:
# Call Python REPL
repl_python()We can then interact with Python session as we would typically do, as if R did not exist.
What is more, we can reach R objects, created in our R session.
All such objects are accessible from within r object in Python session.
To return back into R session we can execute command exit or quit.
Then, in our R session we would be able to reach previously created Python objects as parts of py object.
A quick example:
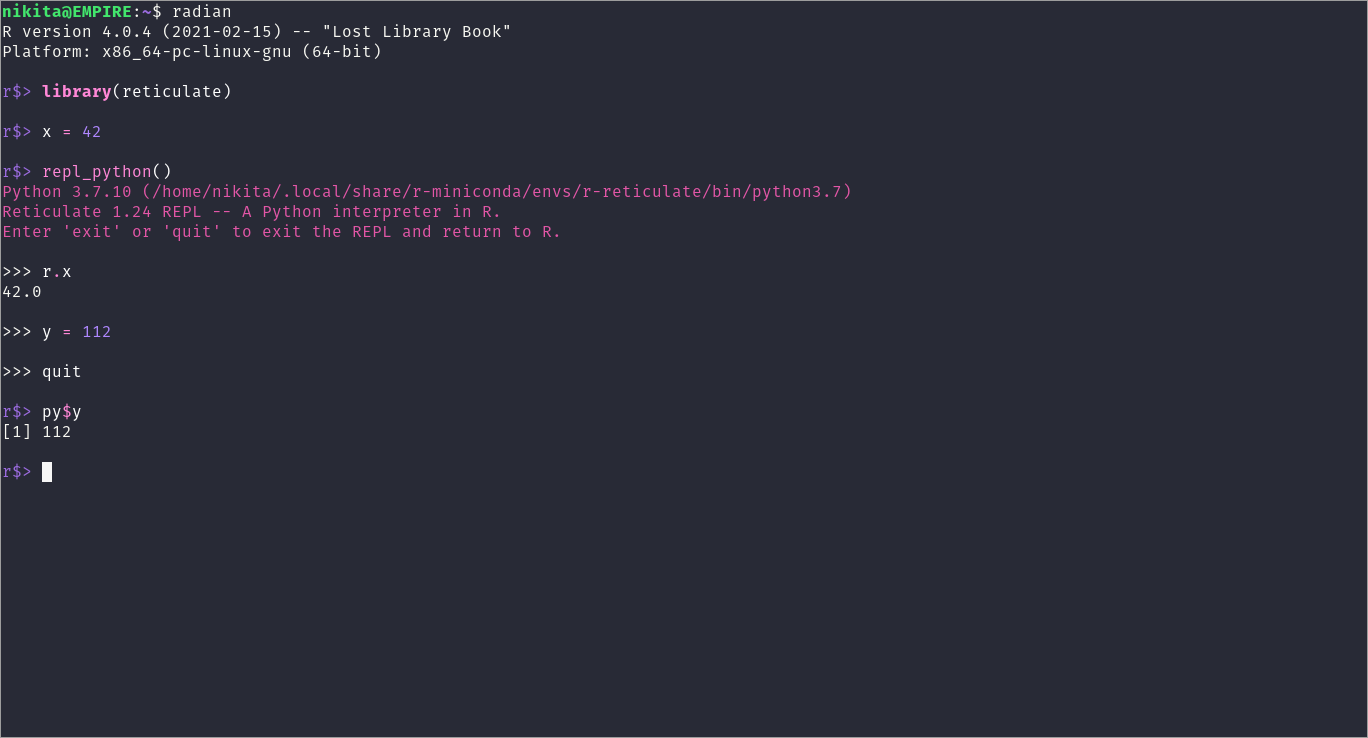
REPL Python example
Now it becomes more clear how the different interfaces interact through dummy objects py and r
Importing Python modules into R
Another option, if you do not want to switch between different languages and interpreters is to load (or import) Python modules inside R.
This can be done via import() function.
# Import numpy
os = import("os")
# List directory contents with Python toolset
os$listdir(".")## [1] "script.py" "index.Rmd" "index.html" "index.Rmd.lock~"
## [5] "index_files"As one can see, the Python libraries are imported into R as separate objects with desired functionalities inside.
Sourcing Python code from files
The same principles may be used with scripts. Meaning you can write the desired Python script for importing data or session setup into a standalone file and then source it from within your R session.
For example, we can create a short script creating a new variable, and save into a file:
# Write into file
cat(
"import os\nld = os.listdir('.')",
append = FALSE,
file = "script.py"
)And then call it from within our R session, and print results:
# Source Python script
source_python("script.py")
# Results
py$ld## [1] "script.py" "index.Rmd" "index.html" "index.Rmd.lock~"
## [5] "index_files"Python chunks inside .Rmd
The final option is brought to us alongside rmarkdown format functionality.
In particular, we can simply create entire Python chunks inside .Rmd file.
It remains only to specify the correct language in chunk’s parameters:
```python
import os
os.listdir(".")
```
```
## ['script.py', 'index.Rmd', 'index.html', 'index.Rmd.lock~', 'index_files']
```The general idea behind this method is common to all other methods. We can access the objects created within Python chunks in R chunks and vice versa:
# Import os
import os
# Check directory contents
dc = os.listdir(".")Then we can access the results from within R:
# Print results
py$dc## [1] "script.py" "index.Rmd" "index.html" "index.Rmd.lock~"
## [5] "index_files"